Cancer stem cells: progress and challenges in lung cancer
Cancer stem cell (CSC) hypothesis
CSC hypothesis suggests cancer is a hierarchically heterogeneous cell population that is dominated and sustained by a very small subpopulation of cancer cells with the property of self-renewal and differentiation. CSCs are thought to be responsible for cancer initiation, progression, metastasis, recurrence, and drug resistance. CSCs were not isolated until the mid-1990s, however, the concept dates back more than one and a half centuries ago. Rudolph Virchow and Julius Cohnheim both German pathologists keenly noted histological similarities between developing fetuses and certain cancers such as teratomas (1). This led them to propose that cancer cells did not spontaneously occur, but rather were the result of activation of embryonic tissue remnants. Formal proof of this concept would have to wait for further technological advancements. In 1963, Bruce et al. (2) observed that only a very small subpopulation of lymphoma cells could form in vitro colonies and initiate tumorgenesis in a xenograft transplant. The first compelling evidence proving the existence of CSCs came in 1997 when Bonnet and Dick (3) isolated a subpopulation of CD34+CD38– acute myeloid leukemia (AML) cells capable of initiating hematopoietic malignancy in mice as well as possessed the capacity to self-renew, proliferate, and differentiate. Since then, proposed CSCs have been isolated from the brain (4), head and neck (5), breast (6), lung (7), liver (8), colon (9), pancreas (10), ovary (11) and prostate (12). Currently, these cells are referred to as “CSCs”, “cancer stem-like cells” (CSLCs), or “tumor-initiating cells” (TICs).
However, the intricacy of cancer demands that the CSC hypothesis be a dynamic hypothesis that must continually be refined as research progresses. Current studies are based on a model in which using surface biomarkers or enzymatic activity, a rare sub-population of cells are isolated from an existing tumor and tested for their ability to form tumor spheroids in vitro and tumors in in vivo through serial xenograft transplantation. The CSC hypothesis, however, has come under scrutiny and remains controversial. For example, critics have challenged whether tumor growth must be initiated by a rare CSC population. Kelly et al. (13) have demonstrated that when lymphomas and leukemias of mouse origin are transplanted into histocompatible mice, a very high frequency of tumor cells (1 in 10) can seed new tumor growth. These findings are suggestive of the limitations of our current xenograft transplantation assay in which the low frequency of CSCs observed may be due in large part to the inability of human tumor cells to adapt to growth in a foreign species microenvironment. Additionally, skeptics of the hypothesis criticize the use of surface biomarkers or enzymatic activity for the identification of CSCs and their lack of utility in therapeutic exploitation. The validity of their claim is substantiated by both the heterogeneity of the proposed CSC markers as well as in the recent discovery of the dynamic state of marker expression on CSCs. The markers that have been identified are not specific to CSC populations, but rather are also abundantly expressed on healthy tissue. Additionally, as detailed in the following review, numerous studies have identified proposed CSC populations which only transiently express the CSC markers. These challenges obfuscate the potential use of biomarkers for antibody- or ligand-targeted therapy in CSCs. Despite this obvious counter indication for use in therapeutic development, there is a grave need to identify and isolate tumor initiating cells in order to further elucidate their complex cellular biology. Regardless of whether CSCs all stably or transiently express biomarkers; have a common or diametric origin; are a rare or abundant population, a greater insight into their mechanism of development, function, and interplay with the tumor microenvironment (TME) will likely result in the development of novel molecular targeting strategies.
Stem cell niches
The stem cell niche is a specialized microenvironment where normal cells and CSCs reside. Microenvironments are defined as the sum total of cell-cell, -extracellular matrix (ECM), and -soluble factor interactions, and the physical states and geometric constraints that a cell may experience (14). The niche is composed of a heterogeneous population of cells that provide normal stem cells with signals to proliferate, differentiate, and undergo asymmetrical cellular division (15). Thus, normal stem cells are regulated by extrinsic cytokines from the niche as well as by intrinsic genetic programs within the stem cell (16). The niche must be pliable to coordinate both homeostasis and repair; however, such flexibility can be heisted by chronic diseases and cancer.
It is unclear whether CSCs direct the formation of the TME or take advantage of the resident microenvironment. The solid TME is composed of a multitude of cell types including fibroblasts and myofibroblasts, immune cells, and mesenchymal stem cells (MSCs) (17). Activated fibroblasts (CAFs) constitute the majority of stromal cells and regulate a variety of pro-tumorigenic processes through their production of various growth factors, chemokines, extracellular proteins, and metalloproteinases (18-21). Strikingly, the switch from a polarized epithelial to a non-polarized mesenchymal cell type with stem cell properties the so called epithelial-to-mesenchymal transition (EMT) shows remarkable similarities to processes that take place during embryonic development (22). In this model, reciprocity between cancer cells and signals emanating from the microenvironment such as growth factors and hypoxia may cause aberrant activation of developmental signaling pathways. Activation of such pathways may induce EMT that imparts self-renewal and multipotent potential of more differentiated tumor cells to CSCs generating heterogeneous tumor cell populations (Figure 1).
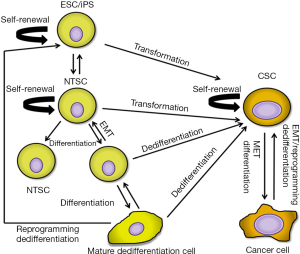
A number of processes are thought to contribute to the establishment of a TME. One such process is chronic inflammation from wounding or carcinogen exposure. Chronic carcinogen exposure, such as cigarette smoke, is the leading cause of lung cancer (23). Researchers have proven that inflammation from chronic chemical irritant exposure enhances cell proliferation and activates the microenvironment (24). Recently, researchers have shown that following lung injury release of local signaling factors [i.e., Platelet Derived Growth Factor-BB (PDGF-BB), Wingless-type MMTV integration site (Wnt), and Transforming Growth Factor-beta (TGF-β)] to lung MSCs facilitates their differentiation into myofibroblasts for cellular repair (25). It is plausible that uncontrolled manipulation of these factors results in deregulated cellular remodeling and a source of maintenance and differentiation signaling for CSCs.
Li et al. (15) proposed that one of the differences between normal stem cells and CSCs is their degree of dependence on the stem cell niche. In general it is clear that there is crosstalk between CSCs and the TME. Hypoxia, inflammation, and desmoplasia influence CSCs through secretion of factors that activate developmental signaling pathways. Conversely, CSCs secrete pro-angiogenic and growth factors to promote a tumor-supportive TME. These studies show that elements of the TME are instructive to tumor cells making them more or less tumorigenic. Collectively, the niche may provide CSCs with dynamic features like growth factors and cytokines that provide cues for niche-induced transformation/dedifferentiation suggesting that molecular targeting of the TME may represent an adjuvant therapeutic strategy for targeting CSCs.
Lung stem cells
The precise origin of CSCs is currently a matter of great conjecture. The most supported hypothesis suggests CSCs arise from normal tissue-specific stem cells in their tissues of origin (26,27). Complicating the identification of normal lung stem cells, lung epithelium is normally thought to be in a quiescent state. In this regard researchers have turned to observing in mice cellular response to lung injury to find cells with stem-like characteristics, such as self-renewal and multipotent potential (28-38). In mice, lung stem cells have been found to exist in functionally and anatomically distinct locations including branch points of airway tubes and junctions between the conducting airway and gas exchange regions. Current data supports that problems with stem cells directly contributes to disease development and progression. Therefore, the following section will highlight our current understanding of normal lung cells capable of stem-like behavior, their cellular response to injury, and their possible pathological association to lung cancer development.
Proximal airway (trachea and bronchi)
The proximal airway consists of the pharynx, larynx, trachea and bronchi. The tracheal epithelium is composed mainly of columnar and mucus secreting goblet cells that function to trap and clear foreign particles as well as lubricate the proximal airway. Cellular turnover in the adult trachea is very low (39) until epithelial injury elicits a rapid response of proliferation in surviving cells, except ciliated cells (40), to repair the tissue. Schoch et al. (41) conducted the initial studies in purification and characterization of airway epithelial stem cells. A subset of mouse tracheal epithelial basal cells located in positions purported to be stem cell niches identified by high keratin 5 promoter activity displayed the stemness capacity to form colonies. Following these studies, rat tracheal basal cells were found to be competent in reconstituting a complete mucociliary epithelium in denuded tracheal grafts (42). Significantly, adult and fetal human tracheal basal cells in a similar model were also capable of differentiating and proliferating to form a fully differentiated mucociliary and functional airway epithelium (43,44). Human bronchial basal cells have also been shown by in vitro assays to form heterogeneous spheres further supporting that human basal cells are capable of both self-renewal and differentiation (29).
Human lung and esophageal squamous cell carcinoma (SCC) are both commonly associated with amplification of chromosomal segment 3q26.33. Curiously, this locus also contains the transcription factor gene Sox2. Sox2 is important in embryological development for normal esophageal squamous development (45), promotes differentiation and proliferation of basal tracheal cells (46), and participates in induction of pluripotent stem cells (47-49). These findings support the role of Sox2 as a lineage survival oncogene in basal cells resulting in SCC.
Midlevel airway (bronchioles)
Nonciliated Clara cells function to detoxify and protect bronchiolar epithelium. Almost four decades ago they were first suggested as stem/progenitor cells when following oxidant induced damage they were capable of self-renewal and differentiation into ciliated cells (50). Nowadays, Clara cells are identified by the biomarker Clara Cell Secretory Protein (CCSP). The inability to easily isolate Clara cells from tissue samples has substantially impeded the critical analysis of these cells in vitro. Recently, however, Wang et al. (51) using a novel isolation method characterized murine lung CCSP-expressing cells that exhibited a number of stem cell-like properties including expression of stem cell markers CD44, CD133, and Sox2; bronchosphere colony formation; and self-renewal capacity. The supposition of Clara cells as progenitors for the airway epithelium has been widely disputed due in large part to the identification of subpopulations of CCSP-expressing cells that may or may not be independent cell populations, such as variant Clara cells (52), type A cells (50), OCT4-expressing stem cells (53), and bronchioalveolar stem cells (BASCs) (36). Clara cells are believed to have multipotent capacity for differentiation, but do not possess the ability to self-renew. An exception to this statement are the variant Clara cells, which are associated with neuroepithelial bodies (NEB), express lower levels of CCSP, and are deficient in cytochrome P450, the enzyme needed to metabolize environmental pollutants (54). Both variant Clara cells and nearby pulmonary neuroendocrine cells (PNECs) have been shown to proliferate and participate in regeneration of rodent epithelium following treatment with the secretory cell specific toxin naphthalene. Moreover, activation of PNECs in the absence of variant Clara cells resulted in PNEC self-renewal without differentiation resulting in hyperplasia but an inability to undergo full epithelial regeneration (34,54,55).
While the specific cell of origin that gives rise to small cell lung carcinoma (SCLC) remains to be elucidated, it is alluring that SCLC predominately localizes to the midlevel bronchioles and is associated like PNECs with primitive neuroendocrine features, such as expression of the neuropeptide Calcitonin-Gene Related Peptide (CGRP) (54), potentiating that PNECs may be the origin of SCLC.
Distal airway (alveoli)
The distal airways include the terminal bronchioles and alveoli and function in gas exchange. BASCs have been identified at the bronchioalveolar duct junction (BADJ) and are characterized by their expression of both CCSP and type II cell markers (SPC) initially suggesting that BASCs were stem cells for both bronchiolar and alveolar epithelium (36,56). Later studies using lineage labeled BASCs demonstrated that these cells contributed to maintenance and repair of the lung airway (bronchiolar), but not alveolar epithelium (57). Recently, scientists have discovered in a murine model distal airway stem cells (DASCs) expressing p63 that generated alveoli in vitro and in vivo following lung injury induced by infection (58). Importantly, conditional expression of oncogenic K-ras in murine lungs resulted in aberrant BASCs outgrowth contributing to the formation of atypical adenomatous hyperplasia, a precursor lesion to adenocarcinoma (59). Furthermore, analysis of human lung adenocarcinoma tissue samples has revealed a BASCs phenotype in 52 of 57 cases characterized by expression of SPC, CCSP, and OCT4 (60). Taken together, these studies strongly implicate self-renewing BASCs in the development of murine adeno- and bronchioalveolar carcinomas; however, it remains to be elucidated in human lungs.
Human lung stem cells
Until recently, resident lung stem/progenitor cells had only been unequivocally identified in the lungs of mice. Kajstura et al. (61) presented the first unequivocal evidence in adult human lungs the existence of multipotent resident lung stem cells that could induce lung repair following injury. This distinct subpopulation of cells enriched for by the cell surface marker c-Kit had the ability for self-renewal, clonogenicity, and multipotency. One of their most astonishing findings demonstrated in vivo that these cells after application to severely damaged xenograft lung tissue could give rise to novel airway structures and vasculature successfully rebuilding the complete lung architecture. Additionally, this subpopulation of cells expressed four genes (NANOG, OCT4, SOX2 and KLF4) known to govern the pluripotency of human embryonic stem cells (ESCs).
The presence of a bona fide human lung stem cell does not depreciate the importance of Basal cells, variant Clara cells, PNECs, or BASCs in response to lung repair and regeneration following lung injury or their possible role in lung cancer development. The above monumental study provides strong evidence to support tissue specific stem cells may arise from dedifferentiated normal somatic cells. Researchers have successfully reprogrammed normal adult fibroblasts into induced pluripotent stem (iPS) cells by retrovirus transduction of the four “Yamanaka factors” (Oct4, c-myc, Sox2 and Klf4) (47). It is plausible that the c-Kit positive human lung stem cells are adult somatic lung cells that have been induced or derepressed to dedifferentiate to form a novel committed progeny, which is a phenomenon that has been shown in other organs (62-64). Interestingly, iPS cells, even without the use of a retrovirus, have demonstrated a degree of risk in developing teratomas (65). Taken together, it is rational to speculate that CSCs may arise by aberrant iPS-like cell reprogramming.
Collectively, these studies reveal that lung CSCs may arise not only from tissue-specific normal stem cells, but also from dedifferentiated progenitor cells, mature cells, or cancer cells through a reprogramming-like process (Figure 1). Thus, CSCs likely have more dynamic properties than originally thought by which to replenish at any time from diverse cells in and around the cancer.
Lung stem cell markers
Biomarkers have been widely used in the field of stem cell research to identify and isolate normal tissue stem cells. Recently, the existence and identification of CSCs has been further supported through the use of such biomarkers including CD133, CD90, and CD44. These markers are widely accepted in CSC research for isolation of human CSCs in multiple solid tumors (5,6,9). Despite the accelerated pace of CSC research in other solid tumors, identification and validation of lung CSCs has largely been obstructed by the complexity of the disease and a deficiency in understanding the hierarchical structure of lung epithelial stem cells. Here we will briefly summarize our current understanding of lung CSC markers.
CD133
CD133 (Prom1), a cell surface glycoprotein, was first identified as a useful marker in the selection of hematopoietic and neural stem cells (5,9). Eramo et al. (7) found a rare undifferentiated cell population expressing CD133 in both NSCLC and SCLC specimens. These cells grew indefinitely as tumor spheres, generated tumor xenografts phenotypically identical to the parental tumors, and were resistant to conventional chemotherapy. Other groups confirmed these findings as well as demonstrated both in vivo and in vitro that long-term chemotherapy exposure could enrich for CD133+ cells in lung cancer (66,67). Moreover, other studies demonstrated these cells had an increase in expression of the ESC transcription factor OCT4 (68) and promoted vasculogenesis (69). Importantly, the significance of CD133 expression as a prognostic marker in NSCLC has been controversial (66,67,70-74). Mizugaki et al. (72) observed that CD133 expression was significantly correlated in NSCLC with pathological stages II, III and IV and was an independent factor for poor prognosis. Conversely, Salnikov et al. (74) reported CD133 was indicative of a resistant phenotype and may be used as a predictor for efficacy of cytotoxic therapy, but that it did not represent a prognostic marker for positive in NSCLC. Moreover, other lines of evidence have surfaced to suggest that the original significance of CD133 expression may have been overstated. For example, recently, CD133-negative cells from lung tumors have shown a higher tumorigenic potential (75,76), resistance to chemotherapy (75), and ability to self-renew (76). Cui et al. (77) using both NSCLC and SCLC lines were able to demonstrate that CD133 expression was transiently expressed in all cell lines tested except one SCLC line, H446. These cells had a tendency to stably express CD133 suggesting the controversies arising from the previous studies could be due in large part to the instability of CD133 (77) or possibly to the dynamic state of CSCs. Moreover, critical analysis of CD133 protein chemistry has highlighted the importance of cautiously evaluating the utility of it as a CSC marker. The antibodies used routinely for purification of CD133 positive cells target poorly characterized, discordantly expressed glycosylated epitopes (78). Taken together, these factors and an incomplete understanding of its biological role complicate the use of CD133 expression as a pan-lung CSC marker.
CD44
CD44, a multifunctional transmembrane glycoprotein (79), is commonly expressed in embryonic (79), hematopoietic (80), mesenchymal (81), and certain epithelial CSCs (5,6,10,12,82-86). Physiologically and pathophysiologically CD44 binds hyaluronic acid, an abundant polysaccharide in stem cell niches (87), to facilitate adhesion, differentiation, homing, and migration within normal and CSC niches. In lung cancer models, elevated CD44 expression was initially detected on specific differentiation phenotypes, including activated type II pneumocytes, squamous metaplasia, and NSCLC cells suggesting it may play a role in the progression of disease (88). Leung et al. (89) demonstrated a subpopulation of CD44+ NSCLC cells were cisplatin-resistant and capable of spheroid body formation, in vivo tumor initiation, and serial tumor transplantability as well as expressed the pluripotency genes OCT4, NANOG and SOX2. Moreover, the transplant initiated xenograft tumors were composed of a heterogeneous population of cells similar to those of the parental H1299 cell line supporting in vivo differentiation.
Collectively, CD44 is poised to be a key player in identifying CSCs due to its innate ability to regulate adhesion, differentiation, homing, and migration. Additional in-depth understanding in CD44 signaling pathway for tumor initiation, maintenance, and metastasis as well as its prognostic importance will aide in resolving its applicability as a lung CSC marker.
Aldehyde dehydrogenase (ALDH)
ALDH detoxifies cells by oxidizing intracellular aldehydes and is known to play a role in differentiation of normal stem cells (90). Analysis of patient samples revealed two aldehyde dehydrogenase isozymes, ALDH1A1 and ALDH3A1, are overexpressed in NSCLC cells, atypical pneumocytes, and normal pneumocytes with chronic carcinogen exposure (91). Sullivan et al. (92) further advanced the ALDH association by demonstrating an ALDH+ subpopulation from multiple NSCLC cell lines had enhanced tumorigenicity, clonogenicity, and Notch signaling dependent self-renewal capacity. Similarly, Akunuru et al. (93) demonstrated that an ALDH subpopulation (ALDHhigh) of NSCLC cells had stem cell-like qualities including increased spheroid formation and metastatic activity. Additionally, these cells had a significant increase in expression of the developmental gene Notch1, self-renewal genes hes1 and shh, and EMT markers Twist and Snail. Unlike CD133 and CD44, ALDH is useful in tumor staging and prognosis (92). NSCLC patients harboring tumor cells overexpressing ALDH1A1 have significant resistance to EGFR tyrosine kinase inhibitors and chemotherapy drugs (94) likely resulting in their poor clinical outcome (92).
To date results for ALDH as a stem cell-like marker in lung cancer appear complicated. ALDH enzymatic activity has been demonstrated in both SCLC and NSCLC cell lines in multiple studies (91,95-98). However, analysis of protein expression of the various isozymes in tissue sections of surgical specimens has revealed only association in NSCLC and not SCLC patients (91). Interestingly, other studies (99) have demonstrated ALDH1/2 expression was considerably higher in primary NSCLC cancers than in cell lines, but the expression levels were similar between the two in SCLCs. Moreover, in vivo non-cancerous airway epithelium exhibited high levels of ALDH1/2 expression, whilst in vitro two immortalized non-cancerous airway epithelial cell lines showed very weak expression (99). Taken together, these studies support that ALDH1/2 expression and its significance likely differ with respect to environmental insult, location within the lung, and pathohistological type. These findings imply ALDH expression and activity is likely not useful as a single pan-lung stem cell marker. However, understanding its mechanism of stemness by investigation of the Notch and Sonic Hedgehog (SHh) signaling pathways in enriched ALDH subpopulations may yield advancements in understanding lung CSC development and maintenance.
Other lung CSC markers
CD166, also known as Activated Leukocyte Cell Adhesion Molecule (ALCAM), is involved in angiogenesis, differentiation, homing, and maintenance of hematopoietic stem cells. It is a validated stem cell marker in normal hematopoietic and MSCs (100) as well as widely accepted as a CSC marker in colorectal cancer (101), prostate (102,103), and glioblastoma (104). Recently, Zhang et al. (105) identified from NSCLC patient tumor tissues a subpopulation of CSCs that robustly expressed CD166 compared to all other surface markers examined. CD166+ cells in vivo had the capacity for self-renewal and to initiate primary and secondary xenograft tumors that phenotypically mimicked the parental patient tumor. In vitro these cells formed self-renewing spheres. Excitingly, in immunodeficient mice as few as 1-5 single cells from dissociated primary lung spheres were capable of initiating in vivo tumorigenesis. Knockdown of CD166 expression in the CSCs did not significantly affect tumorigenicity demonstrating CD166 is an inert surface marker capable of enriching for lung CSCs.
Reprogrammed CD166+ lung epithelial cells (LECs) have also been shown to have a primary role in repair in an acute lung injury model. Soh et al. (106) recently reported the ability to efficiently differentiate human ESCs and iPS toward the lung lineage using a defined step-wise differentiation process. The ES/iPS- derived LEC functional capacity to rescue mice from acute lung injury was unequivocally specific to CD166+ differentiated LECs. Further studies are needed to investigate the therapeutic benefit and pathophysiological consequences of CD166 expression. Moreover, it’s potential for enriching CSCs in SCLC patient samples as well as NSCLC and SCLC cell lines remains to be elucidated.
BMI1 polycomb ring finger oncogene (BMI-1) is involved in axial patterning, haematopoiesis, and cell cycle regulation (107). Additionally, it is known to have a critical role in regulating self-renewal and maintenance of normal lung epithelial and BASCs (108) and CSCs (109). Interestingly, it is overexpressed in a stem-cell like subpopulation of NSCLC cells that are highly tumorigenic and resistant to cisplatin therapy (110). Vrzalikova et al. (111) demonstrated BMI1-negative patients who had received adjuvant therapy had a better disease-free survival for stage I and II NSCLC than BMI-1 positive patients. It is noteworthy that in epithelial cells BMI1 represses the tumor suppressor phosphatase and tensin homolog (PTEN) and induces EMT (112). More importantly, knockdown of BMI-1 in a NSCLC model resulted in restoration of PTEN expression and inhibition of VEGF expression (113).
Podocalyxin-like protein 1 (PODXL-1) is a glycosylated sialomucin and known marker of embryonic, mesenchymal, and hematopoietic stem cells and has been described in multiple human malignancies (114-116). Koch et al. (117) found PODXL-1 and BMI-1 were ubiquitously expressed in SCLC due to aberrant epigenetic changes. From these findings they concluded that an ostensible histogenetic link may exist between developing proximal bronchi and SCLCs supporting the role of PODXL-1 as a potential CSC marker in SCLC.
Urokinase plasminogen activator (uPA) and its receptor uPAR play an integral role in regulating pathways important in cell migration and invasion (118). The importance of uPAR signaling was highlighted with the discovery of its role in inhibition of apoptosis (118). Previously, a subpopulation of SCLC cells from multiple cell lines demonstrated multidrug resistance, clonogenicity, and co-expressed the putative CSC marker CD44 (119). Qiu et al. (120) using a SCLC cell line investigated the stem cell-like properties of enriched uPAR+ cells and found these cells were capable of tumor initiation and differentiation, which further supports a role for uPAR in identification of CSCs in SCLC. Based on these findings, future studies should focus on the staging and prognostic value of PODXL-1, BMI-1, and uPAR for SCLC as well as their collective utility as a panel of CSC markers for SCLC.
Much ambiguity remains in distinguishing lung CSC markers. Concomitantly, intratumoral variation in CSC markers strongly suggests that phenotypic markers may not be a reliable identification tool, but rather cellular state may play an important role in defining a CSC. As reviewed by Visvader and Lindeman (121) CSC pool populations are highly heterogeneous between tumors and within individual tumors; additionally, distinct subsets of CSCs within a tumor have been shown to have the potential to interchange phenotypes. The intratumoral heterogeneity may be a result of clonal evolution giving rise to a second set of CSCs, the CSC phenotype may be unstable resulting in reversion of cell surface markers, and/or long-lived dormant CSCs may have been activated in response to self-feedback regulation or a changing stem cell niche. To date in preclinical models most anti-CSC therapies have targeted only one subtype of CSC resulting in an attenuation rather than eradication of the solid tumors. The existence of multiple CSC pools within individual tumors necessitates development of robust marker strategies that consider the dynamic state of CSCs. Furthermore, technological advancements in tumor monitoring are needed in order to identify residual cells that might drive relapse. A deeper understanding of CSC origin, function, and interaction with the microenvironment will aide in profiling the optimum combination of tumor specific CSC markers as well as aide in the identification of residual CSCs.
Targeting pathways in CSCs
Several pathways including the Wnt, Notch, Hedgehog (Hh) and PTEN pathways, which are tightly regulated during embryogenesis, play a key role in the survival, proliferation, and differentiation of normal stem cells and somatic precursor cells. Interestingly, these pathways are aberrantly regulated in cancer and even to a greater degree in a defined subset of cancer precursor cells resulting in abnormal transformation and possibly inciting tumorigenesis (122,123). Figure 2 depicts the regulation and interplay of these signaling pathways.
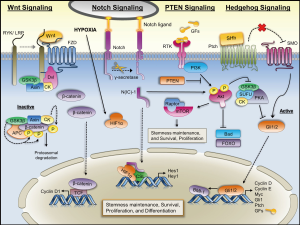
Wnt signaling pathway
The Wnt signaling pathway has been implicated in the control of maintenance, proliferation, and differentiation in both normal stem cells (124-126) and cancer cells (126-128). Wnt binds its receptor Frizzled activating Dvl proteins. These proteins in turn block the Axin/APC/GSK-3β complex derepressing β-catenin facilitating its promotion of cell proliferation by regulating cyclin D1 (123). Mutations in the β-catenin gene, however, are uncommon in lung cancer (129-131). Based on these studies, it appears β-catenin itself plays less of a role in lung carcinogenesis. Previous studies found in NSCLC cell lines and primary cancer tissues Wnt-1 and Wnt-2 were overexpressed and that inhibition of Wnt-1/-2 signaling inhibited cell growth and induced apoptosis (132,133). Similar findings were observed with respect to Dvl-3 and its targeted inhibition (134). Teng et al. (135) demonstrated that cisplatin-resistant A549 cells treated with a GSK-3β inhibitor had an increase in activity of Wnt signaling and expression of OCT4. Strikingly, inhibition of Wnt/β-catenin signaling resulted in a dramatic reduction in OCT4 expression and cisplatin-sensitization. Taken together, these studies provide strong evidence that Wnt signaling plays an important role in lung CSC properties.
Notch signaling pathway
The Notch pathway is an intercellular communication system between a transmembrane ligand and receptor that transduces cell fate regulatory signals (136). There are four known Notch isoforms that appear to have non-overlapping and even opposing effects (136-139). These contextual antagonist roles of Notch signaling in embryonic development can either maintain progenitor/stem cell characteristics or induce tissue-specific differentiation. These diametric roles of Notch pathway activation can possibly explain its tumor suppressive and oncogenic effects. For example, in NSCLC Notch oncogenic activity has most commonly been reported in the context of Notch3 upregulation, but a gain-of-function Notch1 mutation has also been reported (140-142). Other groups have shown that Notch activity facilitates NSCLC survival through inhibition of proapoptotic Bim and induction of antiapoptotic Survivin (143,144). Additionally, constitutively active Notch1/2 in SCLC lines resulted in tumor suppressive effects, whilst a NSCLC ALDHHigh subpopulation has been shown to be dependent on Notch activity for proliferation and clonogenicity (145-147).
Hypoxia induced undifferentiated cell state requires Notch signaling in stem and precursor cell populations (148). Specifically, hypoxia increases expression of genes downstream of Notch and induces hypoxia inducible factor-1 alpha (HIF-1α), a global regulator of oxygen homeostasis, to interact with the intracellular domain of Notch. Chen et al. (149) observed Notch signaling, as measured by γ-secretase cleavage product N(IC)-1, is active in human lung tumor tissue samples and hypoxic lung tumor cell lines, but not normoxic tumor cell lines. Subsequent studies revealed that survival of the lung adenocarcinoma cells during hypoxia was likely mediated by the Insulin Growth Factor-1 receptor (IGF-1R) pathway and subsequent activation of Akt and inhibition of PTEN (150). Collectively, these studies demonstrate the complexity of Notch signaling in lung carcinogenesis and that both cellular (NSCLC vs. SCLC) and microenvironmental (hypoxia) contexts play a profound role in tumor cell response to Notch activation.
Shh signaling pathway
The Shh signaling pathway orchestrates embryonic development as well as regulates stem cell maintenance, cell differentiation, and cell proliferation. Hh proteins bind Patched-1 (PTCH) receptor derepressing the membrane protein Smoothened (SMO). Activation of SMO actuates Gli and its subsequent downstream target Suppressor of Fused Homolog (SUFU) (151). Activation of the Shh pathway has been shown in lung cancer and other cancer models to result in tumorigenesis (152). Watkins et al. (153) revealed that SCLC cells were dependent on Hh signaling for their malignant behavior and underwent autocrine elaboration and reception of the Shh signal within the epithelial compartment similarly to normal airway epithelium during embryonic neuroendocrine differentiation and airway repair. Currently, the mechanism of Hh signaling is not completely understood. However, what is clear is that aberrant Hh signaling does result in tumor growth, proliferation, aggression, and metastasis. Feasibly, other oncogenic factors, such as BMI1, which has been shown to mediate Shh-activated mammosphere formation, may also play a role in CSC self-renewal and differentiation (154).
PTEN signaling pathway
PTEN, a protein tyrosine phosphatase, is a known tumor suppressor gene (155). The observation that loss of PTEN causes or contributes to tumorigenesis in multiple tissues suggests PTEN may play a role in self-renewal and differentiation of TICs (155,156). In fact, PTEN deletion increases stem and progenitor cell populations prior to tumor formation in multiple models including lung adenocarcinoma (157). Additional studies have shown that PTEN plays a pivotal role in lung stem cell homeostasis and cell differentiation (158). PTEN mutations occur at a low frequency in both SCLC and NSCLC. However, in lung cancer other mechanisms to diminish PTEN function or circumscribe PTEN function may be more important. For example, physiological dephosphorylation of its substrate phosphatidylinositol 3,4,5 tri-phosphate (PIP3) results in regulation of a milieu of signaling pathways, such as cell growth, apoptosis, cell cycle progression, DNA damage responses, angiogenesis, and cell polarity (159). Previous reports have shown that 24% of early NSCLC samples had a loss of PTEN expression, which correlated with PTEN promoter methylation (160). Later studies demonstrated that 74% of lung tumors had reduced or lost PTEN expression associated with alterations in TP53 staining (161). As reviewed by Hill and Wu (162), recent studies have revealed a molecular mechanism by which aberrant PTEN expression begets cellular accumulation of PIP3 and resultant activation of Akt pathway, inducing mammalian target of rapamycin (mTOR) and inhibiting forkhead box O (FOXO) and Bcl-2-associated death protein (BAD). Taken together, these studies support a role for PTEN in induction of lung carcinogenesis possibly through deregulation of lung CSCs.
CSC targeted therapies
Standard chemotherapeutic strategies against cancer are severely limited and routinely unsuccessful. Therapeutic failure is commonly associated in multiple malignancies with resistance to chemotherapy and radiotherapy. Additionally, strategies that are initially successful are often times only temporarily curative with recurrence occurring at a later time and routinely in a new site. The CSC hypothesis accounts for these observed patterns of recurrence and metastasis. CSCs are known to be more resistant to conventional chemo- and radio-therapy compared to non-CSC populations. The mechanisms by which CSCs are resistant to conventional therapies is very diverse confounding an already difficult situation. Some of these mechanisms include slowing cell cycle kinetics, efficiently employing mechanisms of DNA repair, intrinsic expression of anti-apoptotic proteins, resistance to oxidative or DNA damage, residence in hypoxic niches, and upregulation of expression of multidrug resistance type membrane transporters. These strategies allow the CSCs to circumvent the mode of action of most all conventional therapies. Clearly, novel therapeutic strategies must be developed to eliminate CSCs and dramatically improve positive patient outcomes and disease-free survival in cancer patients.
The CSC hypothesis has generated a lot of excitement for the discovery and development of therapeutics targeted towards the CSCs. Successful pre-clinical and clinical trial studies have employed strategies of targeting CSCs via use of CSC surface markers, inhibition of developmental stem cell pathways, and ablation of CSC niches. For example, in pre-clinical studies the EGFR antagonist nimotuzumab targeted a glioblastoma subpopulation of cells expressing CD133 rendering them radiosensitive (163). Remarkably, this study also demonstrated CSC markers Nestin and CD133 are co-expressed alongside EGFR. Clinically, the anti-EGFR antibody in combination with conventional therapy has shown promising results in patients with epithelial cancers (164-166) conceivably by radiosensitizing the CSCs. Targeting developmental pathways has also shown clinical success. Von Hoff et al. (167) demonstrated in a cohort of advanced basal cell carcinoma patients a favorable response to treatment with a small-molecule inhibitor of the Hh pathway. Additional studies have investigated other pathways including the Notch and Wnt signaling pathways in multiple solid tumor patient populations (168). Stem cell specific niche constituents and their cognate receptors such as fibronectin and fibronectin receptor in and AML (169) and breast (170), respectively, have also been targeted and demonstrated strong anti-tumorigenic activity.
Turning specifically to lung cancer, researchers have utilized similar strategies in vitro to target lung CSCs. Combinatorial treatment of enriched CD133+/CD44+ lung stem cells with the antipsychotic drug trifluoperazine and gefitinib or cisplatin resulted in a downregulation of CSC-associated gene signatures, inhibition of Wnt signaling, and chemosensitization (171). Therapeutic targeting of the Notch (172) and Wnt pathways (132) has resulted in apoptosis of NSCLCs in vivo and in vitro. Additionally, in vitro targeting of the Hh pathway has shown effective anti-tumorigenic activity in adenocarcinoma and SCLC as well as enhanced the effects of cisplatin (173). CSCs produce Stem Cell Factor (SCF) that binds and activates c-Kit facilitating CSC proliferation. Imatinib inhibits CSC proliferation by blocking SCF. Levina et al. (174) demonstrated that only a small fraction of NSCLC cells express c-Kit rendering imatinib ineffective in a heterogeneous NSCLC population. However, the recent unequivocal identification of human primary resident normal lung stem cells expressing c-Kit (61) conjures the question of imatinib’s effectiveness and utility in eliminating lung CSCs and possible utility as an adjuvant therapeutic.
Several phase I and II studies targeting CSC signaling pathways in solid tumors are underway (175). Pointedly in lung cancer R04929097, a gamma secretase inhibitor, is currently under evaluation. In this study, researchers are interested in the change in expression of Notch pathway and stem cell markers and correlation of this information to therapeutic response (clinicaltrials.gov, NCT01193868). Similar studies with OMP-21M18, a humanized monoclonal antibody against the Notch ligand delta-like 4, are also under investigation (clinicaltrials.gov, NCT01189968). Alternatively, telomerase, which is required for CSC immortalization, has become an attractive CSC target (176). Phase II studies will ascertain if CSC signaling is inhibited by mithramycin, a telomerase inhibitor, in thoracic malignancies (clinicaltrials.gov, NCT01624090). Current, phase I and II studies targeting CSC molecular pathways in lung cancer patients are summarized in Table 1.

Full table
Conclusions and perspective
The CSC model asserts that tumor initiation, metastasis, and recurrence may be driven by a subpopulation of tumor cells resistant to current therapeutic strategies and with the capacity to self-renew and differentiate. The acquisition of self-renewal has led researchers to investigate the possibility of stem cells or stem-like cells as a likely origin of tumorigenesis. However, at present, no experiments have been conducted to definitively prove the existence of CSCs in non-manipulated solid tumors. Historically, bone marrow derived stem cells were identified using radiolabelled cell tracing techniques, which are no longer ethically acceptable in humans. To this extent, the xenograft transplantation assay was developed as a surrogate for defining human CSC. However, there is no direct evidence that the ability of a cell to colonize a foreign microenvironment reflects its hierarchical position within the original tumor. Collectively, these challenges demonstrate the critical need to develop technology for the identification of in situ CSCs as well as more clearly define the xenograft transplantation assay.
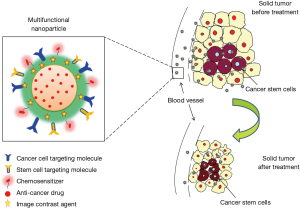
Mounting data supports the existence of CSCs in lung cancer and have been the foundation for developing CSC molecular targeted therapies. In the last several years, researchers have devoted a tremendous amount of resources to the development of novel therapeutic strategies including the innovation of nanomedicine. These therapies can be used to specifically target cell-surface markers, moieties within a stem cell niche, or various signaling pathways as well as used for tumor monitoring. Quintessentially, a multifunctional nanoparticle could be elegantly designed to carry four key elements: (I) targeting moieties; (II) a chemosensitizer; (III) a cytotoxic anticancer drug(s); and (IV) a molecular imaging agent (Figure 3). The ability of the nanoparticle to carry multiple ligands and multiple drugs allows it to be useful against multiple members of a heterogeneous tumor cell population such as tumor cells and CSCs. The provided schematic in Figure 3 is a generic representation of a rudimentary therapeutic strategy. Acquisition of definitive characteristics of the heterogeneous population of tumor cells will allow these therapies to be further refined and adapted for the development of “intelligent nanomedicine.” For example, by understanding the presence or absence of different enzymes in the heterogeneous population, a nanoparticle could be engineered to carry multiple pro-fluorophores that dependent on its enzymatic cleavage would emit a particular color. This color scheme would allow a physician to analyze whether the particle had been taken up by a CSC, normal stem cell, or tumor cell. Such imaging would greatly facilitate the novel application of cell-specific population density tumor monitoring. “Intelligent nanomedicine” would also greatly aide in surgical resection of solid tumors to delineate tumor margins. Additionally photothermal properties could be applied to obliterate remaining tumor cells. With that said, nanoparticles are not necessarily the silver bullet for cancer therapy and are riddled with challenges such as size, charge, biodistribution, and variability in manufacturing, but the conceptual possibilities are limitless.
Regardless, “intelligent nanomedicine” in lung cancer is only theoretical until further investigations can elucidate the hierarchical structure and cell biology of lung cancer, and specifically, lung CSCs. It is imperative that future studies focus on identification of distinguishing properties, including surface markers, gene signatures, anatomical location, and importance of their interactions with their microenvironment. Furthermore, distinguishing CSCs lineage of origin and mechanism(s) of self-renewal will undoubtedly provide substantial insight for targeted therapeutic design and the field of regenerative medicine. And lastly, it will be necessary to gain a deeper insight into the roles self-renewal and maintenance pathways play in normal stem cells and CSCs to accurately identify the best molecular targets for therapy. It is anticipated improved targeted lung CSC therapies will be developed with a greater understanding of CSC biology and advancements in technology.
Acknowledgements
The authors would like to offer our sincerest appreciation to all members of our laboratory for thought provoking discussions and insight.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer 2005;5:311-21. [PubMed]
- Bruce WR, Van Der Gaag H. A Quantitative Assay for the Number of Murine Lymphoma Cells Capable of Proliferation in Vivo. Nature 1963;199:79-80. [PubMed]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730-7. [PubMed]
- Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003;63:5821-8. [PubMed]
- Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A 2007;104:973-8. [PubMed]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100:3983-8. [PubMed]
- Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 2008;15:504-14. [PubMed]
- Yang ZF, Ho DW, Ng MN, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 2008;13:153-66. [PubMed]
- O'Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007;445:106-10. [PubMed]
- Li C, Lee CJ, Simeone DM. Identification of human pancreatic cancer stem cells. Methods Mol Biol 2009;568:161-73. [PubMed]
- Zhang S, Balch C, Chan MW, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res 2008;68:4311-20. [PubMed]
- Collins AT, Berry PA, Hyde C, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005;65:10946-51. [PubMed]
- Kelly PN, Dakic A, Adams JM, et al. Tumor growth need not be driven by rare cancer stem cells. Science 2007;317:337. [PubMed]
- LaBarge MA. The difficulty of targeting cancer stem cell niches. Clin Cancer Res 2010;16:3121-9. [PubMed]
- Li L, Neaves WB. Normal stem cells and cancer stem cells: the niche matters. Cancer Res 2006;66:4553-7. [PubMed]
- Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol 2005;21:605-31. [PubMed]
- Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nature Reviews Cancer 2009;9:239-52. [PubMed]
- Elkabets M, Gifford AM, Scheel C, et al. Human tumors instigate granulin-expressing hematopoietic cells that promote malignancy by activating stromal fibroblasts in mice. J Clin Invest 2011;121:784-99. [PubMed]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature Reviews Cancer 2006;6:392-401. [PubMed]
- Erez N, Truitt M, Olson P, et al. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell 2010;17:135-47. [PubMed]
- Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005;121:335-48. [PubMed]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. [PubMed]
- Doll SR. Smoking and lung cancer. Am J Respir Crit Care Med 2000;162:4-6. [PubMed]
- Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol 2006;1:119-50. [PubMed]
- Foronjy RF, Majka SM. The Potential for Resident Lung Mesenchymal Stem Cells to Promote Functional Tissue Regeneration: Understanding Microenvironmental Cues. Cells 2012;1:874. [PubMed]
- Martínez-Climent JA, Andreu EJ, Prosper F. Somatic stem cells and the origin of cancer. Clin Transl Oncol 2006;8:647-63. [PubMed]
- Pathak S. Organ- and tissue-specific stem cells and carcinogenesis. Anticancer Res 2002;22:1353-6. [PubMed]
- Evans MJ, Van Winkle LS, Fanucchi MV, et al. Cellular and molecular characteristics of basal cells in airway epithelium. Exp Lung Res 2001;27:401-15. [PubMed]
- Rock JR, Onaitis MW, Rawlins EL, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A 2009;106:12771-5. [PubMed]
- Nakajima M, Kawanami O, Jin E, et al. Immunohistochemical and ultrastructural studies of basal cells, Clara cells and bronchiolar cuboidal cells in normal human airways. Pathol Int 1998;48:944-53. [PubMed]
- Giangreco A, Groot KR, Janes SM. Lung cancer and lung stem cells: strange bedfellows? Am J Respir Crit Care Med 2007;175:547-53. [PubMed]
- Boers JE, Ambergen AW, Thunnissen FB. Number and proliferation of basal and parabasal cells in normal human airway epithelium. Am J Respir Crit Care Med 1998;157:2000-6. [PubMed]
- Buckpitt A, Chang AM, Weir A, et al. Relationship of cytochrome P450 activity to Clara cell cytotoxicity. IV. Metabolism of naphthalene and naphthalene oxide in microdissected airways from mice, rats, and hamsters. Mol Pharmacol 1995;47:74-81. [PubMed]
- Stevens TP, McBride JT, Peake JL, et al. Cell proliferation contributes to PNEC hyperplasia after acute airway injury. Am J Physiol 1997;272:L486-93. [PubMed]
- Hong KU, Reynolds SD, Watkins S, et al. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol 2004;286:L643-9. [PubMed]
- Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 2005;121:823-35. [PubMed]
- Stripp BR, Maxson K, Mera R, et al. Plasticity of airway cell proliferation and gene expression after acute naphthalene injury. Am J Physiol 1995;269:L791-9. [PubMed]
- Sullivan JP, Minna JD, Shay JW. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev 2010;29:61-72. [PubMed]
- Kauffman SL. Cell proliferation in the mammalian lung. Int Rev Exp Pathol 1980;22:131-91. [PubMed]
- Rawlins EL, Ostrowski LE, Randell SH, et al. Lung development and repair: contribution of the ciliated lineage. Proc Natl Acad Sci U S A 2007;104:410-7. [PubMed]
- Schoch KG, Lori A, Burns KA, et al. A subset of mouse tracheal epithelial basal cells generates large colonies in vitro. Am J Physiol Lung Cell Mol Physiol 2004;286:L631-42. [PubMed]
- Randell SH, Comment CE, Ramaekers FC, et al. Properties of rat tracheal epithelial cells separated based on expression of cell surface alpha-galactosyl end groups. Am J Respir Cell Mol Biol 1991;4:544-54. [PubMed]
- Hajj R, Baranek T, Le Naour R, et al. Basal cells of the human adult airway surface epithelium retain transit-amplifying cell properties. Stem Cells 2007;25:139-48. [PubMed]
- Avril-Delplanque A, Casal I, Castillon N, et al. Aquaporin-3 expression in human fetal airway epithelial progenitor cells. Stem Cells 2005;23:992-1001. [PubMed]
- Que J, Okubo T, Goldenring JR, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 2007;134:2521-31. [PubMed]
- Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development 2009;136:1899-907. [PubMed]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663-76. [PubMed]
- Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917-20. [PubMed]
- Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007;448:318-24. [PubMed]
- Evans MJ, Johnson LV, Stephens RJ, et al. Renewal of the terminal bronchiolar epithelium in the rat following exposure to NO2 or O3. Lab Invest 1976;35:246-57. [PubMed]
- Wang XY, Keefe KM, Jensen-Taubman SM, et al. Novel method for isolation of murine clara cell secretory protein-expressing cells with traces of stemness. PLoS One 2012;7:e43008. [PubMed]
- Hong KU, Reynolds SD, Giangreco A, et al. Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol 2001;24:671-81. [PubMed]
- Ling TY, Kuo MD, Li CL, et al. Identification of pulmonary Oct-4+ stem/progenitor cells and demonstration of their susceptibility to SARS coronavirus (SARS-CoV) infection in vitro. Proc Natl Acad Sci U S A 2006;103:9530-5. [PubMed]
- Reynolds SD, Giangreco A, Power JH, et al. Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am J Pathol 2000;156:269-78. [PubMed]
- Reynolds SD, Hong KU, Giangreco A, et al. Conditional clara cell ablation reveals a self-renewing progenitor function of pulmonary neuroendocrine cells. Am J Physiol Lung Cell Mol Physiol 2000;278:L1256-63. [PubMed]
- Giangreco A, Reynolds SD, Stripp BR. Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am J Pathol 2002;161:173-82. [PubMed]
- Rawlins EL, Okubo T, Xue Y, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 2009;4:525-34. [PubMed]
- Kumar PA, Hu Y, Yamamoto Y, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 2011;147:525-38. [PubMed]
- Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15:3243-8. [PubMed]
- Zhang XY, Zheng BQ, Han BH, et al. Lung adenocarcinoma stem cell phenotypes and their correlation with patient prognosis. Zhonghua Zhong Liu Za Zhi 2009;31:836-40. [PubMed]
- Kajstura J, Rota M, Hall SR, et al. Evidence for human lung stem cells. N Engl J Med 2011;364:1795-806. [PubMed]
- Chen X, Mao Z, Liu S, et al. Dedifferentiation of adult human myoblasts induced by ciliary neurotrophic factor in vitro. Mol Biol Cell 2005;16:3140-51. [PubMed]
- Porat S, Dor Y. New sources of pancreatic beta cells. Current Diabetes Reports 2007;7:304-8. [PubMed]
- Zhou Q, Brown J, Kanarek A, et al. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 2008;455:627-32. [PubMed]
- Okita K, Nakagawa M, Hyenjong H, et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008;322:949-53. [PubMed]
- Woo T, Okudela K, Mitsui H, et al. Prognostic value of CD133 expression in stage I lung adenocarcinomas. Int J Clin Exp Pathol 2010;4:32-42. [PubMed]
- Zhang HZ, Wei YP, Wang M, et al. Association of CD133 and endothelin-converting enzyme expressions with prognosis of non-small cell lung carcinoma. Nan Fang Yi Ke Da Xue Xue Bao 2007;27:696-9. [PubMed]
- Chen YC, Hsu HS, Chen YW, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One 2008;3:e2637. [PubMed]
- Hilbe W, Dirnhofer S, Oberwasserlechner F, et al. CD133 positive endothelial progenitor cells contribute to the tumour vasculature in non-small cell lung cancer. J Clin Pathol 2004;57:965-9. [PubMed]
- Mihatsch J, Toulany M, Bareiss PM, et al. Selection of radioresistant tumor cells and presence of ALDH1 activity in vitro. Radiother Oncol 2011;99:300-6. [PubMed]
- Li F, Zeng H, Ying K. The combination of stem cell markers CD133 and ABCG2 predicts relapse in stage I non-small cell lung carcinomas. Med Oncol 2011;28:1458-62. [PubMed]
- Mizugaki H, Sakakibara-Konishi J, Kikuchi J, et al. CD133 expression: a potential prognostic marker for non-small cell lung cancers. Int J Clin Oncol 2014;19:254-9. [PubMed]
- Okudela K, Woo T, Mitsui H, et al. Expression of the potential cancer stem cell markers, CD133, CD44, ALDH1, and beta-catenin, in primary lung adenocarcinoma--their prognostic significance. Pathol Int 2012;62:792-801. [PubMed]
- Salnikov AV, Gladkich J, Moldenhauer G, et al. CD133 is indicative for a resistance phenotype but does not represent a prognostic marker for survival of non-small cell lung cancer patients. Int J Cancer 2010;126:950-8. [PubMed]
- Kubo T, Takigawa N, Osawa M, et al. Subpopulation of small-cell lung cancer cells expressing CD133 and CD87 show resistance to chemotherapy. Cancer Sci 2013;104:78-84. [PubMed]
- Meng X, Li M, Wang X, et al. Both CD133+ and CD133- subpopulations of A549 and H446 cells contain cancer-initiating cells. Cancer Sci 2009;100:1040-6. [PubMed]
- Cui F, Wang J, Chen D, et al. CD133 is a temporary marker of cancer stem cells in small cell lung cancer, but not in non-small cell lung cancer. Oncol Rep 2011;25:701-8. [PubMed]
- Bidlingmaier S, Zhu X, Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J Mol Med (Berl) 2008;86:1025-32. [PubMed]
- Haegel H, Dierich A, Ceredig R. CD44 in differentiated embryonic stem cells: surface expression and transcripts encoding multiple variants. Dev Immunol 1994;3:239-46. [PubMed]
- Bruns I, Cadeddu RP, Brueckmann I, et al. Multiple myeloma-related deregulation of bone marrow-derived CD34(+) hematopoietic stem and progenitor cells. Blood 2012;120:2620-30. [PubMed]
- Campagnoli C, Roberts IA, Kumar S, et al. Identification of mesenchymal stem/progenitor cells in human first-trimester fetal blood, liver, and bone marrow. Blood 2001;98:2396-402. [PubMed]
- Du L, Wang H, He L, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res 2008;14:6751-60. [PubMed]
- Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A 2007;104:10158-63. [PubMed]
- Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005;65:5506-11. [PubMed]
- Patrawala L, Calhoun T, Schneider-Broussard R, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene 2006;25:1696-708. [PubMed]
- Rybak AP1, He L, Kapoor A, et al. Characterization of sphere-propagating cells with stem-like properties from DU145 prostate cancer cells. Biochim Biophys Acta 2011;1813:683-94.
- Solis MA, Chen YH, Wong TY, et al. Hyaluronan regulates cell behavior: a potential niche matrix for stem cells. Biochem Res Int 2012;2012:346972.
- Penno MB, August JT, Baylin SB, et al. Expression of CD44 in human lung tumors. Cancer Res 1994;54:1381-7. [PubMed]
- Leung EL, Fiscus RR, Tung JW, et al. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One 2010;5:e14062. [PubMed]
- Russo JE, Hilton J. Characterization of cytosolic aldehyde dehydrogenase from cyclophosphamide resistant L1210 cells. Cancer Res 1988;48:2963-8. [PubMed]
- Patel M, Lu L, Zander DS, et al. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer 2008;59:340-9. [PubMed]
- Sullivan JP, Spinola M, Dodge M, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res 2010;70:9937-48. [PubMed]
- Akunuru S, James Zhai Q, Zheng Y. Non-small cell lung cancer stem/progenitor cells are enriched in multiple distinct phenotypic subpopulations and exhibit plasticity. Cell Death Dis 2012;3:e352. [PubMed]
- Huang CP, Tsai MF, Chang TH, et al. ALDH-positive lung cancer stem cells confer resistance to epidermal growth factor receptor tyrosine kinase inhibitors. Cancer Lett 2013;328:144-51. [PubMed]
- Moreb JS, Baker HV, Chang LJ, et al. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol Cancer 2008;7:87. [PubMed]
- Moreb JS, Zucali JR, Ostmark B, et al. Heterogeneity of aldehyde dehydrogenase expression in lung cancer cell lines is revealed by Aldefluor flow cytometry-based assay. Cytometry B Clin Cytom 2007;72:281-9. [PubMed]
- Moreb JS. Aldehyde dehydrogenase as a marker for stem cells. Curr Stem Cell Res Ther 2008;3:237-46. [PubMed]
- Ucar D, Cogle CR, Zucali JR, et al. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem Biol Interact 2009;178:48-55. [PubMed]
- Okudela K, Nagahara N, Katayama A, et al. Cancer Stem Cells in Lung Cancer: Distinct Differences between Small Cell and Non-Small Cell Lung Carcinomas. In: Shostak S. ed. Cancer Stem Cells Theories and Practice. InTech; 2011.
- Cortés F, Deschaseaux F, Uchida N, et al. HCA, an immunoglobulin-like adhesion molecule present on the earliest human hematopoietic precursor cells, is also expressed by stromal cells in blood-forming tissues. Blood 1999;93:826-37. [PubMed]
- Levin TG, Powell AE, Davies PS, et al. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 2010;139:2072-82.e5.
- Jiao J, Hindoyan A, Wang S, et al. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One 2012;7:e42564. [PubMed]
- Kristiansen G, Pilarsky C, Wissmann C, et al. ALCAM/CD166 is up-regulated in low-grade prostate cancer and progressively lost in high-grade lesions. Prostate 2003;54:34-43. [PubMed]
- Kijima N, Hosen N, Kagawa N, et al. CD166/activated leukocyte cell adhesion molecule is expressed on glioblastoma progenitor cells and involved in the regulation of tumor cell invasion. Neuro Oncol 2012;14:1254-64. [PubMed]
- Zhang WC, Shyh-Chang N, Yang H, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012;148:259-72. [PubMed]
- Soh BS, Zheng D, Li Yeo JS, et al. CD166(pos) subpopulation from differentiated human ES and iPS cells support repair of acute lung injury. Mol Ther 2012;20:2335-46. [PubMed]
- van Kemenade FJ, Raaphorst FM, Blokzijl T, et al. Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood 2001;97:3896-901. [PubMed]
- Zacharek SJ, Fillmore CM, Lau AN, et al. Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell 2011;9:272-81. [PubMed]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 2003;423:255-60. [PubMed]
- Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A 2009;106:16281-6. [PubMed]
- Vrzalikova K, Skarda J, Ehrmann J, et al. Prognostic value of Bmi-1 oncoprotein expression in NSCLC patients: a tissue microarray study. J Cancer Res Clin Oncol 2008;134:1037-42. [PubMed]
- Song LB, Li J, Liao WT, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest 2009;119:3626-36. [PubMed]
- Meng X, Wang Y, Zheng X, et al. shRNA-mediated knockdown of Bmi-1 inhibit lung adenocarcinoma cell migration and metastasis. Lung Cancer 2012;77:24-30. [PubMed]
- Ney JT, Zhou H, Sipos B, et al. Podocalyxin-like protein 1 expression is useful to differentiate pancreatic ductal adenocarcinomas from adenocarcinomas of the biliary and gastrointestinal tracts. Hum Pathol 2007;38:359-64. [PubMed]
- Somasiri A, Nielsen JS, Makretsov N, et al. Overexpression of the anti-adhesin podocalyxin is an independent predictor of breast cancer progression. Cancer Res 2004;64:5068-73. [PubMed]
- Kelley TW, Huntsman D, McNagny KM, et al. Podocalyxin: a marker of blasts in acute leukemia. Am J Clin Pathol 2005;124:134-42. [PubMed]
- Koch LK, Zhou H, Ellinger J, et al. Stem cell marker expression in small cell lung carcinoma and developing lung tissue. Hum Pathol 2008;39:1597-605. [PubMed]
- Alfano D, Franco P, Vocca I, et al. The urokinase plasminogen activator and its receptor: role in cell growth and apoptosis. Thromb Haemost 2005;93:205-11. [PubMed]
- Gutova M, Najbauer J, Gevorgyan A, et al. Identification of uPAR-positive chemoresistant cells in small cell lung cancer. PLoS One 2007;2:e243. [PubMed]
- Qiu X, Wang Z, Li Y, et al. Characterization of sphere-forming cells with stem-like properties from the small cell lung cancer cell line H446. Cancer Lett 2012;323:161-70. [PubMed]
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012;10:717-28. [PubMed]
- Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature 2001;414:105-11. [PubMed]
- Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer 2003;3:895-902. [PubMed]
- Batlle E, Henderson JT, Beghtel H, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 2002;111:251-63. [PubMed]
- Korinek V, Barker N, Moerer P, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 1998;19:379-83. [PubMed]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature 2005;434:843-50. [PubMed]
- van de Wetering M, Sancho E, Verweij C, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002;111:241-50. [PubMed]
- He B, Barg RN, You L, et al. Wnt signaling in stem cells and non-small-cell lung cancer. Clin Lung Cancer 2005;7:54-60. [PubMed]
- Sunaga N, Kohno T, Kolligs FT, et al. Constitutive activation of the Wnt signaling pathway by CTNNB1 (beta-catenin) mutations in a subset of human lung adenocarcinoma. Genes Chromosomes Cancer 2001;30:316-21. [PubMed]
- Shigemitsu K, Sekido Y, Usami N, et al. Genetic alteration of the beta-catenin gene (CTNNB1) in human lung cancer and malignant mesothelioma and identification of a new 3p21.3 homozygous deletion. Oncogene 2001;20:4249-57. [PubMed]
- Ueda M, Gemmill RM, West J, et al. Mutations of the beta- and gamma-catenin genes are uncommon in human lung, breast, kidney, cervical and ovarian carcinomas. Br J Cancer 2001;85:64-8. [PubMed]
- He B, You L, Uematsu K, et al. A monoclonal antibody against Wnt-1 induces apoptosis in human cancer cells. Neoplasia 2004;6:7-14. [PubMed]
- You L, He B, Xu Z, et al. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene 2004;23:6170-4. [PubMed]
- Uematsu K, He B, You L, et al. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene 2003;22:7218-21. [PubMed]
- Teng Y, Wang X, Wang Y, et al. Wnt/beta-catenin signaling regulates cancer stem cells in lung cancer A549 cells. Biochem Biophys Res Commun 2010;392:373-9. [PubMed]
- Pannuti A, Foreman K, Rizzo P, et al. Targeting Notch to target cancer stem cells. Clin Cancer Res 2010;16:3141-52. [PubMed]
- Bigas A, Martin DI, Milner LA. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol Cell Biol 1998;18:2324-33. [PubMed]
- Fan X, Mikolaenko I, Elhassan I, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res 2004;64:7787-93. [PubMed]
- Shimizu K, Chiba S, Saito T. Functional diversity among Notch1, Notch2, and Notch3 receptors. Biochem Biophys Res Commun 2002;291:775-9. [PubMed]
- Westhoff B, Colaluca IN, D’Ario G, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A 2009;106:22293-8. [PubMed]
- Dang TP, Gazdar AF, Virmani AK, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst 2000;92:1355-7. [PubMed]
- Haruki N, Kawaguchi KS, Eichenberger S, et al. Dominant-negative Notch3 receptor inhibits mitogen-activated protein kinase pathway and the growth of human lung cancers. Cancer Res 2005;65:3555-61. [PubMed]
- Konishi J, Yi F, Chen X, et al. Notch3 cooperates with the EGFR pathway to modulate apoptosis through the induction of bim. Oncogene 2010;29:589-96. [PubMed]
- Chen Y, Li D, Liu H, et al. Notch-1 signaling facilitates survivin expression in human non-small cell lung cancer cells. Cancer Biol Ther 2011;11:14-21. [PubMed]
- Sriuranpong V, Borges MW, Ravi RK, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res 2001;61:3200-5. [PubMed]
- Sriuranpong V, Borges MW, Strock CL, et al. Notch signaling induces rapid degradation of achaete-scute homolog 1. Mol Cell Biol 2002;22:3129-39. [PubMed]
- Ball DW. Achaete-scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett 2004;204:159-69. [PubMed]
- Gustafsson MV, Zheng X, Pereira T, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell 2005;9:617-28. [PubMed]
- Chen Y, De Marco MA, Graziani I, et al. Oxygen concentration determines the biological effects of NOTCH-1 signaling in adenocarcinoma of the lung. Cancer Res 2007;67:7954-9. [PubMed]
- Eliasz S, Liang S, Chen Y, et al. Notch-1 stimulates survival of lung adenocarcinoma cells during hypoxia by activating the IGF-1R pathway. Oncogene 2010;29:2488-98. [PubMed]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature 2001;411:349-54. [PubMed]
- Yuan Z, Goetz JA, Singh S, et al. Frequent requirement of hedgehog signaling in non-small cell lung carcinoma. Oncogene 2007;26:1046-55. [PubMed]
- Watkins DN, Berman DM, Burkholder SG, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature 2003;422:313-7. [PubMed]
- Liu S, Dontu G, Mantle ID, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res 2006;66:6063-71. [PubMed]
- Ali IU, Schriml LM, Dean M. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. J Natl Cancer Inst 1999;91:1922-32. [PubMed]
- Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci U S A 1999;96:4240-5. [PubMed]
- Tiozzo C, De Langhe S, Yu M, et al. Deletion of Pten expands lung epithelial progenitor pools and confers resistance to airway injury. Am J Respir Crit Care Med 2009;180:701-12. [PubMed]
- Yanagi S, Kishimoto H, Kawahara K, et al. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J Clin Invest 2007;117:2929-40. [PubMed]
- Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene 2008;27:5477-85. [PubMed]
- Soria JC, Lee HY, Lee JI, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res 2002;8:1178-84. [PubMed]
- Marsit CJ, Zheng S, Aldape K, et al. PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum Pathol 2005;36:768-76. [PubMed]
- Hill R, Wu H. PTEN, stem cells, and cancer stem cells. J Biol Chem 2009;284:11755-9. [PubMed]
- Diaz Miqueli A, Rolff J, Lemm M, et al. Radiosensitisation of U87MG brain tumours by anti-epidermal growth factor receptor monoclonal antibodies. Br J Cancer 2009;100:950-8. [PubMed]
- Crombet T, Osorio M, Cruz T, et al. Use of the humanized anti-epidermal growth factor receptor monoclonal antibody h-R3 in combination with radiotherapy in the treatment of locally advanced head and neck cancer patients. J Clin Oncol 2004;22:1646-54. [PubMed]
- Ramos TC, Figueredo J, Catala M, et al. Treatment of high-grade glioma patients with the humanized anti-epidermal growth factor receptor (EGFR) antibody h-R3: report from a phase I/II trial. Cancer Biol Ther 2006;5:375-9. [PubMed]
- Ramakrishnan MS, Eswaraiah A, Crombet T, et al. Nimotuzumab, a promising therapeutic monoclonal for treatment of tumors of epithelial origin. MAbs 2009;1:41-8. [PubMed]
- Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med 2009;361:1164-72. [PubMed]
- Diaz A, Leon K. Therapeutic Approaches to Target Cancer Stem Cells. Cancers (Basel) 2011;3:3331-52. [PubMed]
- Matsunaga T, Takemoto N, Sato T, et al. Interaction between leukemic-cell VLA-4 and stromal fibronectin is a decisive factor for minimal residual disease of acute myelogenous leukemia. Nat Med 2003;9:1158-65. [PubMed]
- Sandal T, Valyi-Nagy K, Spencer VA, et al. Epigenetic reversion of breast carcinoma phenotype is accompanied by changes in DNA sequestration as measured by AluI restriction enzyme. Am J Pathol 2007;170:1739-49. [PubMed]
- Yeh CT, Wu AT, Chang PM, et al. Trifluoperazine, an antipsychotic agent, inhibits cancer stem cell growth and overcomes drug resistance of lung cancer. Am J Respir Crit Care Med 2012;186:1180-8. [PubMed]
- Konishi J, Kawaguchi KS, Vo H, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res 2007;67:8051-7. [PubMed]
- Tian F, Mysliwietz J, Ellwart J, et al. Effects of the Hedgehog pathway inhibitor GDC-0449 on lung cancer cell lines are mediated by side populations. Clin Exp Med 2012;12:25-30. [PubMed]
- Levina V, Marrangoni A, Wang T, et al. Elimination of human lung cancer stem cells through targeting of the stem cell factor-c-kit autocrine signaling loop. Cancer Res 2010;70:338-46. [PubMed]
- Gottschling S, Schnabel PA, Herth FJ, et al. Are we missing the target? Cancer stem cells and drug resistance in non-small cell lung cancer. Cancer Genomics Proteomics 2012;9:275-86. [PubMed]
- Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer 2008;8:167-79. [PubMed]
Cite this article as: Templeton AK, Miyamoto S, Babu A, Munshi A, Ramesh R. Cancer stem cells: progress and challenges in lung cancer. Stem Cell Investig 2014;1:9.


