Regulation of cancer stem cells by RING finger ubiquitin ligases
Introduction
In the past several decades, cancer stem cells (CSCs, also known as cancer initiating cells) have been intensively investigated. Normal tissue stem cells give rise to transit-amplifying cells or progenitor cells by asymmetric cell division which in turn generates more differentiated cells in a given tissue. Similarly, CSCs are capable of self-renewal, either by symmetric or asymmetric cell division, and are able to reproduce malignant tumor cells indefinitely (1). Studies of the molecular mechanisms governing CSC maintenance and differentiation have mostly focused on the roles of signaling and transcriptional processes. However, it has recently been demonstrated that protein modification via the ubiquitin system is also crucial for the normal and abnormal stem cell functions, and the loss of such modifications can lead to tumorigenesis and progression. Thus, understanding the emerging field of how the ubiquitin modification system regulates CSC activity may lead to novel targeted cancer therapeutics.
Cancer stem cell (CRC)
CSCs constitute a minority population in tumors and have low proliferative rate (2). They may originate from stem cells which have gained cancerous properties through genetic and epigenetic changes. Alternatively, they may arise from transformed progenitor cells that have acquired self-renewal capabilities (1).
CSCs reside in specialized microenvironments called niches, which play an important role in stem cell maintenance. The constituents of niche include fibroblasts, endothelial cells, perivascular cells, tissue macrophages, extracellular matrix, and soluble factors excreted from cells. There is cross talk between CSCs and the niche; CSCs instruct the formation of niche, whereas the niche governs the proliferation, differentiation, invasion and metastasis of CSCs or cancer cells (3). For example, the hypoxic locations in the tumor can function as niche for CSC, and induce stem-like characteristics of tumor cells through hypoxia inducible factor-1 (HIF-1) in many tumors. The stemness induction is achieved by activation of transcription factors involved in reprogramming of induced pleuripotent stem cells (i.e., Oct4, Sox2, Nanog and KLF4) (4).
Current cancer therapies frequently fail to eliminate advanced tumors, which may be due to their inability to effectively target CSC populations. The embryonic pathways such as Wnt, Hedgehog, and Notch control self-renewal and cell fate decisions of stem cells and progenitor cells. And these evolutionary conserved pathways are also involved in CSC maintenance (5). Thus, targeting these pathways or the interactions between CSC and tumor microenvironment may be effective in eradicating CSCs and preventing radiation or chemotherapy resistance.
The ubiquitin system
The attachment of ubiquitin polypeptides to intracellular proteins is a key mechanism in regulating many cellular processes. Ubiquitin is covalently attached to target proteins via an isopeptide bond between its C-terminal glycine and a lysine residue of the acceptor substrate. Assembly of a chain of at least four ubiquitins linked through their Lys48 residue marks cellular proteins for degradation by the 26S proteasome. In contrast, monoubiquitination or polyubiquitination with chains linked via Lys63 serve as nonproteolytic signals for intracellular trafficking, DNA repair, and signal transduction pathways. The ubiquitin modification of proteins occurs through an enzymatic cascade consisting of the ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes (6). E1 enzyme activates ubiquitin in an ATP-dependent manner, and subsequently transfers ubiquitin to E2. The E3 ligase binds to both the substrate and E2 conjugated with ubiquitin, and facilitates the ubiquitin transfer from E2 to substrates. The pairing of E2s and substrates by E3s determines the specificity in ubiquitination.
There are two major types of E3s in eukaryotes, defined by the presence of either a HECT or a RING domain (6). The HECT family comprises large proteins, each with the ability to interact directly with E2 enzymes and their specific substrates. By contrast, RING finger E3 ligases are multi-subunit complexes that bind the E2 ligase via a catalytic ring finger protein and interact with substrates via a separate receptor protein. RING finger ubiquitin ligases, which are the focus of this review, are conserved from yeast to humans, with about 616 different RING finger proteins potentially expressed in human cells (7). However, many aspects of these enzymes remain poorly understood.
It has become evident that many RING finger E3 ligases are implicated in malignancy. Oncogenic transformation is characterized by dysregulated cell growth signals, insensitivity to anti-growth or pro-apoptotic signals, dysregulation of the cell cycle and genomic instability. Solid tumors also acquire the ability to induce angiogenesis and metastasis (8). RING finger proteins are implicated in all of these steps. We will focus below on specific RING finger ubiquitin ligases that regulate different properties of CSCs (Figure 1), which might provide potential therapy targets for the treatment of various types of malignancies.
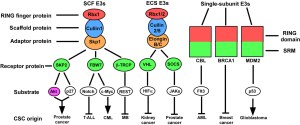
SCF RING finger E3 ligase
The SCF complex represents one of the largest classes of RING finger ubiquitin ligases. It consists of three invariable components: Rbx1 (RING finger protein, binding with E2 enzyme), Cullin1 (scaffold protein), and Skp1 (adaptor protein). It also has one variable component, known as F-box protein, that binds to Skp1 through its F-box motif and recognizes substrates through different substrate-interacting motifs (SIMs) (9). Up to 69 F-box proteins (in humans) can potentially serve as substrate recognition elements. Among them, there are several well-studied F-box proteins involved in the control of cell-cycle progression, and the self-renewal and differentiation of stem cells.
S phase kinase-associated protein 2 (Skp2)
Skp2 is an F-box protein of SCF E3 ligase complex, and recognizes substrates through the leucine-rich repeats (LRR) motif. In addition to forming the canonical SCF E3 complex, Skp2 also binds with ankyrin-repeat SOCS box-containing protein 2 (Asb2), and facilitate the formation of a non-canonical dimeric E3 ligase complexes containing not only Cullin1-Skp1, but also Cullin5-ElonginB/C E3 ligases, which promotes the degradation of substrates of both Cullin1 and Cullin5 (10).
Although Skp2 targets numerous proteins involved in various biological processes, such as cyclins, E2F1, Foxo1 and E47 (9), G1/S cyclin-dependent kinase inhibitor (p27) seems to be the best-studied target of Skp2 (11). p27 is a tumor suppressor, and Skp2 regulates apoptosis, cell-cycle progression, and proliferation through promoting the ubiquitination and degradation of p27 (12). Skp2 knock-out mice are viable, but the cells contain markedly enlarged nuclei with polyploidy and multiple centrosomes, show a reduced growth rate and increased apoptosis. Skp2 deficient cells exhibit increased accumulation of p27 (11). Over-expression of Skp2 is frequently observed in human cancers. The ectopic expression of Skp2 promotes tumorigenesis and metastasis in prostate tumor models (13), and induces T cell lymphomas in mice cooperating with activated N-Ras (14). Furthermore, it is recently reported that Skp2 triggers the nonproteolytic K63-linked ubiquitination of Akt, and promotes Akt-mediated glycolysis and tumorigenesis (15).
The dysregulation of cell-cycle and aerobic glycolysis contribute to the self-renewal and proliferation of CSC. Knock-down and pharmacological inhibition of Skp2 significantly reduce the ALDH+ CSC population of prostate cancer, and down-regulate the sphere formation capability of prostate cancer CSC (16). Thus, targeting Skp2 is a promising strategy for cancer treatment.
F-box and WD repeat-containing protein 7 (Fbw7)
Fbw7 is another F-box protein of the SCF complex that targets several oncoproteins, including cyclin E, c-Myc, and Notch, for ubiquitination and degradation through its WD-40 motif. It is thus thought to function as a tumor suppressor (9). Germ-line deletion of Fbw7 results in embryonic lethality at around E10.5 due to vascular defects, which are attributed to the stabilization of Notch4 in the embryo (17). Conditional deletion of Fbw7 in the hematopoietic system leads to an increase in the frequency of actively cycling LSK (Lin–, c-Kit+, Sca-1+) population, with eventual exhaustion of hematopoietic stem cells (HSCs) (18). Furthermore, Fbw7–/– LSK cells down-regulate genes involved in HSC quiescence, implying a global loss of quiescence characteristics (19).
The regulation of cell-cycle by Fbw7 is mainly through targeting cyclin E and Notch for degradation. Cyclin E is involved in driving cells in the G0 or G1 phase into the S phase and, as predicted, is frequently dysregulated in cancer (20). Notch is important for directing lymphoid lineage cell fate determination and has also been implicated in HSC self-renewal (21,22). Notch is expressed by HSCs while its ligand, Jagged, is expressed by the HSC niche, and increased Jagged/Notch activation results in increased HSC number and niche expansion (23). Notch is a potent oncogene in T-cell acute lymphoblastic leukemia (T-ALL), and the aberrant activation of Notch promotes the self-renewal of leukemia stem cell and drives the leukemic phenotype (24). And over 10% of these malignancies exhibit either a mutation or homozygous deletion of Fbw7 gene. The loss of Fbw7 function promotes the development of T-ALL through stabilization of Notch proteins (25).
In contrast with T-ALL, Fbw7 has an important function for the initiation and the progression of chronic myelogenous leukemia (CML). Deletion of Fbw7 leads to c-Myc over-expression in leukemic initiating cells (LICs). Although c-Myc is able to promote tumor progression, unphysiological over-expression of oncogenic c-Myc inhibits tumor growth through inducing p53-dependent apoptosis. Thus, the ablation of Fbw7 results in the apoptosis in LICs, and eventually the inhibition of CML progression (26).
β-TrCP
F-box and WD repeat-containing protein 1A (β-TrCP) is a versatile F-box protein of the SCF E3 ligase complex that targets various substrates for degradation. β-catenin and IκBα are well-established targets of β-TrCP for ubiquitylation and degradation (27). The role of β-TrCP in tumor progression is paradoxical. On one hand, it promotes the degradation of β-catenin and thus inactivates the Wnt pathway, which stimulates the proliferation of cancer cells (28). The genetic alterations of β-TrCP gene and accumulation of β-catenin have been shown in several studies on human gastric cancer and prostate cancer (29). On the other hand, it also accelerates the turnover of IκBα and activates the NFκB pathway, which antagonizes the pro-apoptotic signals and enhances the chemo-resistance of cancer cells (30).
Recent report indicates that β-TrCP is involved in the normal differentiation of neural stem cell (NSC) through promoting the degradation of a transcriptional repressor REST (31). REST is found to be over-expressed in human medulloblastoma (MB), which represents one of the most malignant brain tumors in children and is believed to arise from undifferentiated NSC present in the cerebellum. The ectopic expression of REST in v-myc-immortalized NSC promotes MB formation in mice (32). Thus, up-regulation of β-TrCP could be a promising therapy for this deadly disease by promoting the terminal differentiation of MB stem cell.
ECS RING finger E3 ligase
ECS complex is another major class of RING finger E3 ligases, and includes three invariable subunits: Elongin B/C (adaptor protein), Cullin2/5 (scaffold protein) and Rbx1/2 (RING finger protein, binding with E2). It also has one variable element for substrate recognition, known as BC-box protein since it binds with Elongin B/C through BC-box domain (33). Among the BC-box proteins there are two major populations, SOCS-box proteins and VHL-box proteins. The SOCS-box containing proteins specifically interact with Cullin5 and Rbx2, whereas the VHL-box proteins form a complex with Cullin2 and Rbx1. SOCS-box protein population are mainly composed of four families, SOCS, WSB, SSB and ASB, which respectively contain SH2 domain, WD-40 repeats, SPRY domain and Ankyrin repeats N-terminal to the SOCS box (34). They target various components of cytokine signaling for degradation, and negatively regulates cellular proliferation signal (10,33-36). While the VHL-box proteins, including von Hippel-Lindau tumour suppressor (VHL), LRR-1 and FEM1B, facilitate the degradation of proteins involved in the stress response and cellular metabolism (33,37).
The suppressor of cytokine signaling (SOCS)
SOCS proteins are well-studied substrate recognition subunits of Cullin5 ECS complex. They recognize the phosphorylated tyrosine residues of substrates through SH2 domains, and promote the ubiquitination and degradation of those proteins. The reported targets of SOCS proteins include the p65 subunit of NFκB, myeloid differentiation primary-response gene 88 (MyD88)-adaptor-like protein (MAL) and Janus protein kinases (JAKs), which play pivotal roles in inflammation, as well as in the progression of cancers (38,39).
JAK/STAT pathways are important intracellular signaling cascades to transduce the differentiation and proliferation signals from cytokines, growth factors, and hormonal factors. Once activated by the receptors of above molecules, JAKs promote the dimerization and activation of downstream transcription factors, such as members of the signal transduction and activators of transcription (STAT) family. STAT dimers then translocate to the nucleus where they bind IFN-γ-activated (GAS)-like elements, leading to the transcriptional activation of multiple genes (40). Persistent JAK/STAT activation is observed in many cancer cells, including colorectal cancer, prostate cancer, breast cancer, and leukemia (41). Consistently, promoter hypermethylation and decreased expression of SOCS1 and SOCS3 are detected in various cancers (42,43). Mice with SOCS3 deletion in gastrointestinal epithelial cell (T3b-SOCS3 cKO mice) display the aberrant JAK-STAT signaling and severe phenotype of gastric cancer (44). According to a recent report, the elevated activation of IL-6/JAK/STAT3 pathway is observed in the stem-like cells of prostate cancer patients, and the abnormal activation promotes the clonogenicity of stem-like cancer cells and the outgrowth of castration-resistant tumor (45). Therefore, up-regulation of SOCS proteins, especially SOCS3 could be a promising treatment for advanced prostate cancer.
VHL
VHL was first discovered as a tumor suppressor gene that is inactivated in the familial kidney cancer syndrome VHL disease (46). Approximately 57% of sporadic clear cell cancers of the kidney contain inactivating mutations of VHL, and 98% of these have loss of heterozygosity (LOH) at the VHL locus (47). As the substrate recognition subunit of Cullin2 ECS complex, VHL targets Hypoxia inducible factor α (HIFα), an important transcription factor promoting cell survival under hypoxic conditions. Under normoxic conditions, VHL recognizes the hydroxylated proline of HIFα protein, and facilitates its degradation (48).
The loss of VHL protein stabilizes HIFα protein and induces HSC quiescence, which is determined by an increase in LSK (Lin–, c-Kit+, Sca-1+) cell number in G0 phase and the attenuated differentiation status in peripheral blood (49). Furthermore, the hypoxia environment and stabilized HIF expand the sub-population of cancer cells positive for CSC markers, and promote a stem-like phenotype in cancer cells. This phenotype of CSC may contribute to the recurrence after radiation or chemotherapy by reducing ROS and enhancing the activity of DNA checkpoint kinases to prevent DNA damage (5). Thus, introduction of VHL to cancer cells could be a promising method to reduce the therapeutic resistance.
Single-subunit RING finger E3 ligase
There are also a large number of RING finger proteins mediating substrate ubiquitination individually. They contain RING finger domain binding to E2 enzymes, and various SIMs, and are involved in different stages of cancer progression.
CBL
Casitas B cell lymphoma (C-Cbl), the cellular homolog of the v-Cbl oncogene, encodes a single-subunit RING finger E3 ligase, and contains a highly conserved N-terminal tyrosine kinase-binding (TKB) domain that mediates interactions between c-Cbl and phosphorylated tyrosine residues on its substrates (50). It can regulate the protein levels of c-Kit, STAT5 and Flt3, all of which contribute to HSC maintenance (51,52).
Cbl knock-out mice exhibit aberrant hematopoiesis. Cbl–/– HSCs show enhanced reconstitution capacity in competitive bone marrow transplantation (BMT) assays and more proliferation in BrdU incorporation experiments. Cbl–/– LSKs have increased levels of phospho-STAT5 and c-Myc mRNA (STAT5 is an activator of Myc transcription) suggesting Cbl deficiency stabilizes active STAT5 and promotes the hyperproliferative phenotype (24).
Cbl is mutated in 5-15% of human myeloid leukemia. Mice containing loss-of-function mutations of the c-Cbl gene develop aggressive myeloid leukemia, and the leukemic stem cells from those mice exhibit augmented signaling from Flt3, which is a receptor tyrosine kinase (RTK) and a substrate of c-Cbl E3 ubiqutin ligase (53). Thus, the ubiquitination and degradation of RTK signaling components by c-Cbl might also contribute to its effect on CSC proliferation.
BRCA1
BRCA1 is a tumor suppressor that is frequently mutated in familial breast and ovarian cancer (54). Similar to c-Cbl, BRCA1 is a single-subunit RING finger E3 ligase, and contains two BRCA1 C-terminal (BRCT) domains, which recognize the phosphorylation sites of substrates (55). The role of BRCA1 in DNA repair was well established. BRCA1 deficiency leads to a defect in the repair of double-stranded breaks by homologous recombination (HR), which is responsible for the genomic instability and tumorigenesis (56). A key ubiquitination substrate of BRCA1 is CTBP interacting protein (CTIP), which is the binding partner of the transcriptional repressor CTBP, and is involved in the checkpoint arrest in response to DNA damage (57).
Recently a role of BRCA1 in stem cell regulation and the control of mammary gland differentiation have been suggested. BRCA1 expression is required for the differentiation of ER-negative stem/progenitor cells to ER-positive mature luminal cells (58). Loss of BRCA1 may result in the accumulation of genetically unstable breast stem cells or luminal progenitor cells, providing prime targets for further transformation.
MDM2
MDM2 is a single subunit RING finger E3 ligase that targets p53 for degradation, as well as inhibits the transcriptional activity of p53 by binding to its N-terminus (59). Loss of MDM2 activity in HSC leads to stabilized p53, which impedes hematopoiesis via induction of cell cycle arrest, senescence and ultimately cell death of HSCs and progenitors (60).
MDM2 acts as an oncoprotein, and is up-regulated in glioblastoma stem cells through MEK-ERK signaling. MDM2 prevents p53 function and maintains the expression of O6-methylguanine DNA methyltransferase (MGMT), which is a key factor in conferring the Temozolomide (TMZ) resistance to glioma stem cells (61).
Targeted therapy towards RING fingers
Since the RING finger E3 ligases play important roles in regulating the self-renewal, differentiation and therapeutic resistance of CSCs, specific inhibition on certain RING finger proteins may provide promising strategies for cancer treatment. RING finger proteins facilitate ubiquitin transfer from E2 directly to the substrate, but they are not catalysts with active sites. Thus, inhibitor development would probably have to be focused on disrupting the RING structure or the RING-E2 interface. Some specific inhibitors of RING finger E3 ligases have been developed. For example, Nutlin-3 is a novel small-molecule antagonist of MDM2 that binds MDM2 in the p53-binding pocket, thereby interfering with MDM2-directed p53 degradation. The p53 stabilization results in apoptosis in cancer cells (62). Another promising chemical inhibitor can bind with Trp97 of Skp2 and prevents the interaction between Skp2 and its adaptor protein Skp1. This inhibitor can effectively reduce the CSC population in prostate cancer, and overcome chemo-resistance (16). Therefore, with the accumulation of structural and functional data and the elucidation of the pathways that are controlled by RING finger E3s, there is great potential to apply this knowledge to the development of novel targeted therapeutics.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Takebe N, Ivy SP. Controversies in cancer stem cells: targeting embryonic signaling pathways. Clin Cancer Res 2010;16:3106-12. [PubMed]
- Reiman JM, Knutson KL, Radisky DC. Immune promotion of epithelial-mesenchymal transition and generation of breast cancer stem cells. Cancer Res 2010;70:3005-8. [PubMed]
- Nguyen LV, Vanner R, Dirks P, et al. Cancer stem cells: an evolving concept. Nat Rev Cancer 2012;12:133-43. [PubMed]
- Heddleston JM, Li Z, Lathia JD, et al. Hypoxia inducible factors in cancer stem cells. Br J Cancer 2010;102:789-95. [PubMed]
- Maugeri-Saccà M, Vigneri P, De Maria R. Cancer stem cells and chemosensitivity. Clin Cancer Res 2011;17:4942-7. [PubMed]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2001;2:169-78. [PubMed]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009;78:399-434. [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [PubMed]
- Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol 2005;16:323-33. [PubMed]
- Nie L, Zhao Y, Wu W, et al. Notch-induced Asb2 expression promotes protein ubiquitination by forming non-canonical E3 ligase complexes. Cell Res 2011;21:754-69. [PubMed]
- Nakayama K, Nagahama H, Minamishima YA, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J 2000;19:2069-81. [PubMed]
- Kossatz U, Dietrich N, Zender L, et al. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev 2004;18:2602-7. [PubMed]
- Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer 2008;112:1415-24. [PubMed]
- Latres E, Chiarle R, Schulman BA, et al. Role of the F-box protein Skp2 in lymphomagenesis. Proc Natl Acad Sci U S A 2001;98:2515-20. [PubMed]
- Chan CH, Li CF, Yang WL, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 2012;149:1098-111. [PubMed]
- Chan CH, Morrow JK, Li CF, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell 2013;154:556-68. [PubMed]
- Tetzlaff MT, Yu W, Li M, et al. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc Natl Acad Sci U S A 2004;101:3338-45. [PubMed]
- Matsuoka S, Oike Y, Onoyama I, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev 2008;22:986-91. [PubMed]
- Thompson BJ, Jankovic V, Gao J, et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med 2008;205:1395-408. [PubMed]
- Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature 1999;401:297-300. [PubMed]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science 1999;284:770-6. [PubMed]
- Nie L, Perry SS, Zhao Y, et al. Regulation of lymphocyte development by cell-type-specific interpretation of Notch signals. Mol Cell Biol 2008;28:2078-90. [PubMed]
- Duncan AW, Rattis FM, DiMascio LN, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol 2005;6:314-22. [PubMed]
- Rathinam C, Thien CB, Langdon WY, et al. The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev 2008;22:992-7. [PubMed]
- O’Neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med 2007;204:1813-24. [PubMed]
- Reavie L, Buckley SM, Loizou E, et al. Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell 2013;23:362-75. [PubMed]
- Nakayama K, Hatakeyama S, Maruyama S, et al. Impaired degradation of inhibitory subunit of NF-kappa B (I kappa B) and beta-catenin as a result of targeted disruption of the beta-TrCP1 gene. Proc Natl Acad Sci U S A 2003;100:8752-7. [PubMed]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 1998;391:493-6. [PubMed]
- Gerstein AV, Almeida TA, Zhao G, et al. APC/CTNNB1 (beta-catenin) pathway alterations in human prostate cancers. Genes Chromosomes Cancer 2002;34:9-16. [PubMed]
- Müerköster S, Arlt A, Sipos B, et al. Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer Res 2005;65:1316-24. [PubMed]
- Westbrook TF, Hu G, Ang XL, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 2008;452:370-4. [PubMed]
- Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle 2006;5:1929-35. [PubMed]
- Kamura T, Maenaka K, Kotoshiba S, et al. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev 2004;18:3055-65. [PubMed]
- Hilton DJ, Richardson RT, Alexander WS, et al. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc Natl Acad Sci U S A 1998;95:114-9. [PubMed]
- Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 2011;11:629-43. [PubMed]
- Lisztwan J, Imbert G, Wirbelauer C, et al. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev 1999;13:1822-33. [PubMed]
- Pozzebon ME, Varadaraj A, Mattoscio D, et al. BC-box protein domain-related mechanism for VHL protein degradation. Proc Natl Acad Sci U S A 2013;110:18168-73. [PubMed]
- Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol 2007;7:454-65. [PubMed]
- Inagaki-Ohara K, Kondo T, Ito M, et al. SOCS, inflammation, and cancer. JAKSTAT 2013;2:e24053. [PubMed]
- O’Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 2002;109 Suppl:S121-31. [PubMed]
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol 2007;7:41-51. [PubMed]
- Pierconti F, Martini M, Pinto F, et al. Epigenetic silencing of SOCS3 identifies a subset of prostate cancer with an aggressive behavior. Prostate 2011;71:318-25. [PubMed]
- Sakamoto LH. MT1G hypermethylation: a potential prognostic marker for hepatoblastoma. Pediatr Res 2010;67:387-93. [PubMed]
- Inagaki-Ohara K, Mayuzumi H, Kato S, et al. Enhancement of leptin receptor signaling by SOCS3 deficiency induces development of gastric tumors in mice. Oncogene 2014;33:74-84. [PubMed]
- Kroon P, Berry PA, Stower MJ, et al. JAK-STAT blockade inhibits tumor initiation and clonogenic recovery of prostate cancer stem-like cells. Cancer Res 2013;73:5288-98. [PubMed]
- Latif F, Tory K, Gnarra J, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 1993;260:1317-20. [PubMed]
- Gnarra JR, Tory K, Weng Y, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 1994;7:85-90. [PubMed]
- Kaelin WG Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer 2008;8:865-73. [PubMed]
- Takubo K, Goda N, Yamada W, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 2010;7:391-402. [PubMed]
- Zheng N, Wang P, Jeffrey PD, et al. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 2000;102:533-9. [PubMed]
- Goh EL, Zhu T, Leong WY, et al. c-Cbl is a negative regulator of GH-stimulated STAT5-mediated transcription. Endocrinology 2002;143:3590-603. [PubMed]
- Zeng S, Xu Z, Lipkowitz S, et al. Regulation of stem cell factor receptor signaling by Cbl family proteins (Cbl-b/c-Cbl). Blood 2005;105:226-32. [PubMed]
- Reindl C, Quentmeier H, Petropoulos K, et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin Cancer Res 2009;15:2238-47. [PubMed]
- Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet 2001;10:705-13. [PubMed]
- Moynahan ME, Chiu JW, Koller BH, et al. Brca1 controls homology-directed DNA repair. Mol Cell 1999;4:511-8. [PubMed]
- Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 2010;11:138-48. [PubMed]
- Yu X, Fu S, Lai M, et al. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev 2006;20:1721-6. [PubMed]
- Buckley NE, Mullan PB. BRCA1--conductor of the breast stem cell orchestra: the role of BRCA1 in mammary gland development and identification of cell of origin of BRCA1 mutant breast cancer. Stem Cell Rev 2012;8:982-93. [PubMed]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997;420:25-7. [PubMed]
- Abbas HA, Maccio DR, Coskun S, et al. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell 2010;7:606-17. [PubMed]
- Sato A, Sunayama J, Matsuda K, et al. MEK-ERK signaling dictates DNA-repair gene MGMT expression and temozolomide resistance of stem-like glioblastoma cells via the MDM2-p53 axis. Stem Cells 2011;29:1942-51. [PubMed]
- Tabe Y, Sebasigari D, Jin L, et al. MDM2 antagonist nutlin-3 displays antiproliferative and proapoptotic activity in mantle cell lymphoma. Clin Cancer Res 2009;15:933-42. [PubMed]
Cite this article as: Kang B, Sun XH. Regulation of cancer stem cells by RING finger ubiquitin ligases. Stem Cell Investig 2014;1:5.


